Common molecular subtypes of liver cancer
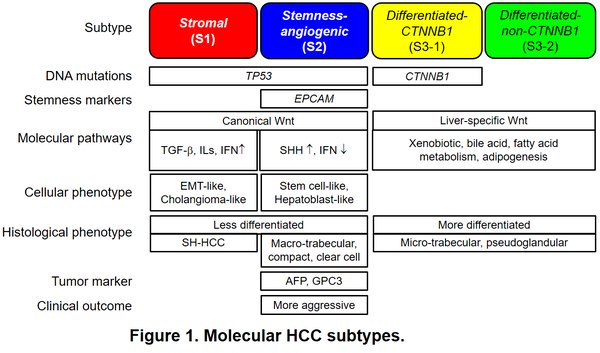
Hepatocellular carcinoma (HCC) is the primary histological type of liver cancer, one of the most lethal cancer types worldwide. HCC tumors are known to be heterogeneous histologically, molecularly, and clinically. Geographic heterogeneity in liver disease etiology (e.g., hepatitis B, hepatitis C, alcohol, fatty liver, and metabolic disorders) and patient race/ethnicity adds additional complexity. In consistent, molecular profiling studies have elucidated highly diverse molecular aberrations in HCC tumors and associated molecular and clinical phenotypes across the regions. To answer the question whether there are commonly observed molecular subtypes of HCC tumors across the global patient populations, we previously performed transcriptome meta-analysis for discovery and validation of molecular subtypes of HCC in 603 patients enrolled from Asia, Europe, and the U.S., encompassing diverse liver disease etiologies and patient races/ethnicities. This analysis successfully identified three major molecular subtypes robustly observed across the patient cohorts as a molecular measure of inter-tumor heterogeneity. As expected, global transcriptome recapitulates various pathogenic genetic/epigenetic alterations and molecular pathway dysregulations characteristic to each subtype (Figure 1).
Hepatocellular carcinoma (HCC) is the primary histological type of liver cancer, one of the most lethal cancer types worldwide. HCC tumors are known to be heterogeneous histologically, molecularly, and clinically. Geographic heterogeneity in liver disease etiology (e.g., hepatitis B, hepatitis C, alcohol, fatty liver, and metabolic disorders) and patient race/ethnicity adds additional complexity. In consistent, molecular profiling studies have elucidated highly diverse molecular aberrations in HCC tumors and associated molecular and clinical phenotypes across the regions. To answer the question whether there are commonly observed molecular subtypes of HCC tumors across the global patient populations, we previously performed transcriptome meta-analysis for discovery and validation of molecular subtypes of HCC in 603 patients enrolled from Asia, Europe, and the U.S., encompassing diverse liver disease etiologies and patient races/ethnicities. This analysis successfully identified three major molecular subtypes robustly observed across the patient cohorts as a molecular measure of inter-tumor heterogeneity. As expected, global transcriptome recapitulates various pathogenic genetic/epigenetic alterations and molecular pathway dysregulations characteristic to each subtype (Figure 1).
HCC tumors can be classified into stromal subtype with fibrotic phenotype and immune cell infiltrates (S1 subtype), stemness-angiogenic subtype with positivity of stemness/tumor markers such as AFP, EPCAM, and GPC3 as well as activation of angiogenic drivers such as VEGFA (S2 subtype), and differentiated subtype with matured hepatocyte-like phenotype (S3), which is further sub-classified according to presence or absence of CTNNB1 gene mutations (S3-1 or S3-2 subtype, respectively). Of note, the subtypes are correlated with microscopic morphological appearance, i.e., histological architectures and cytological features such as micro-trabecular pattern, macro-trabecular/compact pattern, pseudo glandular pattern, clear cell variant, and steatohepatitic variant, often representing intra-tumor heterogeneity in an HCC tumor as the “nodule-in-nodule” structure. This histology-subtype correlation was subsequently confirmed by other researchers. While single-cell-level transcriptomic diversity is noted among tumor and tumor-associated stromal/immune cells, the subtype likely reflect dominant tumor clones and the entire eco system of the tumor nodule, which will provide clues to effective therapeutic intervention. The common HCC subtyping has been widely used as a reference classification in multiple integrative multi-omic molecular profiling studies led by national and international consortia such as The Cancer Genome Atlas (TCGA) consortium.
Subtype-targeted therapy
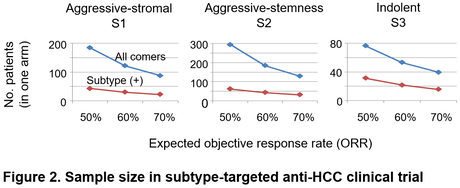
Given the low objective response rates of currently available medical therapies in HCC (only up to 5-30%), it is rational to indicate specific molecular-targeted agents only to a subset of tumors most likely respond to the therapy to maximize its cost-effectiveness and minimize adverse effects in non-responders. As a proof of this concept, efficacy of monoclonal anti-VEGF receptor 2, ramucirumab, was shown to be limited to HCC with elevated serum biomarker, alpha-fetoprotein (AFP), a hallmark of the S2 subtype. Similarly, a TGF-beta receptor 1 inhibitor, galunisertib, showed anti-HCC effect in TGF-beta-activated and AFP-low tumors (suggestively S1 subtype). These clinical observations suggest utility of molecular subtyping of HCC tumors to guide personalized indication of medical therapies. In addition, clinical trials enriched for specific subtype can be conducted with two- to four-fold smaller sample size (Figure 2). Furthermore, the HCC subtypes can be recapitulated in in vitro and in vivo human-material- and animal-based experimental models, in which subtype-specific drug response can be monitored and functionally assessed. Single-cell omics profiling combined with Digital Spatial Profiling (DSP) enables identification of more precise cellular mechanisms of disease progression and drug response.
Subtype-targeted therapy
Given the low objective response rates of currently available medical therapies in HCC (only up to 5-30%), it is rational to indicate specific molecular-targeted agents only to a subset of tumors most likely respond to the therapy to maximize its cost-effectiveness and minimize adverse effects in non-responders. As a proof of this concept, efficacy of monoclonal anti-VEGF receptor 2, ramucirumab, was shown to be limited to HCC with elevated serum biomarker, alpha-fetoprotein (AFP), a hallmark of the S2 subtype. Similarly, a TGF-beta receptor 1 inhibitor, galunisertib, showed anti-HCC effect in TGF-beta-activated and AFP-low tumors (suggestively S1 subtype). These clinical observations suggest utility of molecular subtyping of HCC tumors to guide personalized indication of medical therapies. In addition, clinical trials enriched for specific subtype can be conducted with two- to four-fold smaller sample size (Figure 2). Furthermore, the HCC subtypes can be recapitulated in in vitro and in vivo human-material- and animal-based experimental models, in which subtype-specific drug response can be monitored and functionally assessed. Single-cell omics profiling combined with Digital Spatial Profiling (DSP) enables identification of more precise cellular mechanisms of disease progression and drug response.
References
1. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589-604.
2. Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol. 2015;2(4):371-9.
3. Dhanasekaran R, Nault JC, Roberts LR, Zucman-Rossi J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology. 2019;156(2):492-509.
4. Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385-92.
5. Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667-76.
6. Tan PS, Nakagawa S, Goossens N, Venkatesh A, Huang T, Ward SC et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int. 2016;36(1):108-18.
7. Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouze E, Blanc JF et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67(4):727-38.
8. Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68(3):1025-41.
9. Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71(3):616-30.
10. Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36(4):418-30.
11. Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327-41.
12. Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257-61.
13. Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell. 2017;32(1):57-70.
14. Parikh ND, Singal AG, Hutton DW. Cost effectiveness of regorafenib as second-line therapy for patients with advanced hepatocellular carcinoma. Cancer. 2017;123(19):3725-31.
15. Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(2):282-96.
16. Faivre S, Santoro A, Kelley RK, Gane E, Costentin CE, Gueorguieva I et al. Novel transforming growth factor beta receptor I kinase inhibitor galunisertib (LY2157299) in advanced hepatocellular carcinoma. Liver Int. 2019;39(8):1468-77.
17. Zhu S, Hoshida Y. Molecular heterogeneity in hepatocellular carcinoma. Hepat Oncol. 2018;5(1):HEP10.
18. Hirschfield H, Bian CB, Higashi T, Nakagawa S, Zeleke TZ, Nair VD et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med. 2018;50(1):e419.
19. Schmidt B, Wei L, DePeralta DK, Hoshida Y, Tan PS, Sun X et al. Molecular subclasses of hepatocellular carcinoma predict sensitivity to fibroblast growth factor receptor inhibition. Int J Cancer. 2016;138(6):1494-505.
20. Qiu Z, Li H, Zhang Z, Zhu Z, He S, Wang X et al. A Pharmacogenomic Landscape in Human Liver Cancers. Cancer Cell. 2019;36(2):179-93.
21. Dow M, Pyke RM, Tsui BY, Alexandrov LB, Nakagawa H, Taniguchi K et al. Integrative genomic analysis of mouse and human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2018;115(42):E9879-E88..
Past and current grants
NIH/NCI U01CA226052, Mehta/Drake/Singal/Hoshida (MPI)
CPRIT, RR180016, Hoshida (PI)
Irma T. Hirschl/Monique Weill-Caulier Scholar Award, Hoshida (PI)
Clinical trials
NCT04145141, Hoshida (site PI)
1. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589-604.
2. Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol. 2015;2(4):371-9.
3. Dhanasekaran R, Nault JC, Roberts LR, Zucman-Rossi J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology. 2019;156(2):492-509.
4. Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385-92.
5. Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667-76.
6. Tan PS, Nakagawa S, Goossens N, Venkatesh A, Huang T, Ward SC et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int. 2016;36(1):108-18.
7. Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouze E, Blanc JF et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67(4):727-38.
8. Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68(3):1025-41.
9. Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71(3):616-30.
10. Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019;36(4):418-30.
11. Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327-41.
12. Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257-61.
13. Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell. 2017;32(1):57-70.
14. Parikh ND, Singal AG, Hutton DW. Cost effectiveness of regorafenib as second-line therapy for patients with advanced hepatocellular carcinoma. Cancer. 2017;123(19):3725-31.
15. Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(2):282-96.
16. Faivre S, Santoro A, Kelley RK, Gane E, Costentin CE, Gueorguieva I et al. Novel transforming growth factor beta receptor I kinase inhibitor galunisertib (LY2157299) in advanced hepatocellular carcinoma. Liver Int. 2019;39(8):1468-77.
17. Zhu S, Hoshida Y. Molecular heterogeneity in hepatocellular carcinoma. Hepat Oncol. 2018;5(1):HEP10.
18. Hirschfield H, Bian CB, Higashi T, Nakagawa S, Zeleke TZ, Nair VD et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med. 2018;50(1):e419.
19. Schmidt B, Wei L, DePeralta DK, Hoshida Y, Tan PS, Sun X et al. Molecular subclasses of hepatocellular carcinoma predict sensitivity to fibroblast growth factor receptor inhibition. Int J Cancer. 2016;138(6):1494-505.
20. Qiu Z, Li H, Zhang Z, Zhu Z, He S, Wang X et al. A Pharmacogenomic Landscape in Human Liver Cancers. Cancer Cell. 2019;36(2):179-93.
21. Dow M, Pyke RM, Tsui BY, Alexandrov LB, Nakagawa H, Taniguchi K et al. Integrative genomic analysis of mouse and human hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2018;115(42):E9879-E88..
Past and current grants
NIH/NCI U01CA226052, Mehta/Drake/Singal/Hoshida (MPI)
CPRIT, RR180016, Hoshida (PI)
Irma T. Hirschl/Monique Weill-Caulier Scholar Award, Hoshida (PI)
Clinical trials
NCT04145141, Hoshida (site PI)